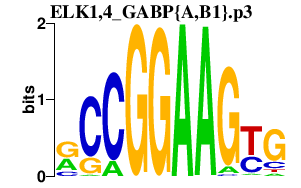
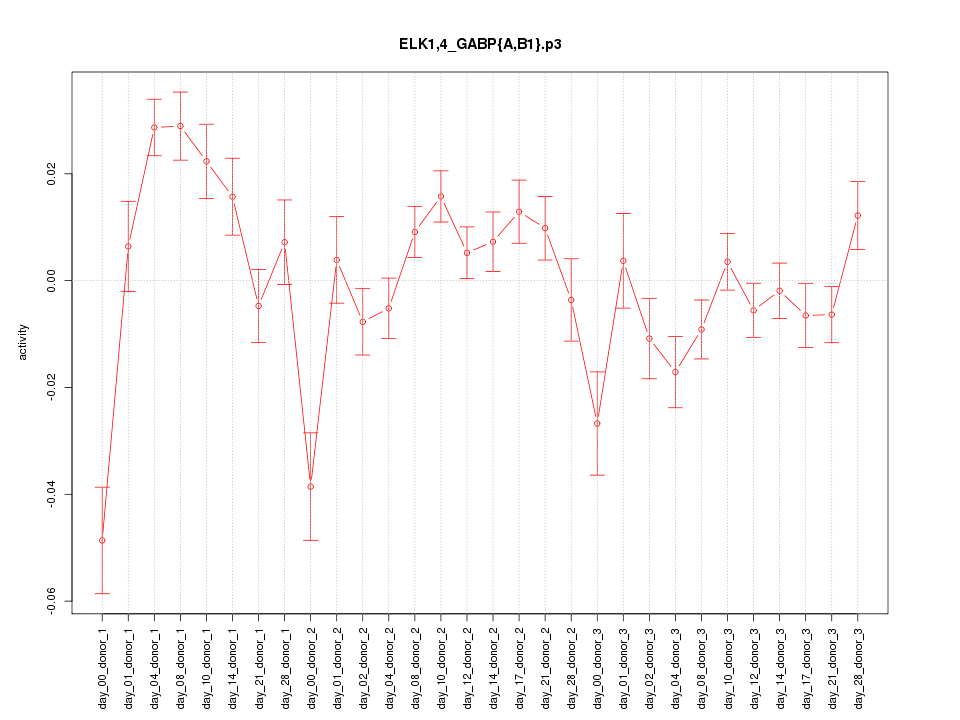
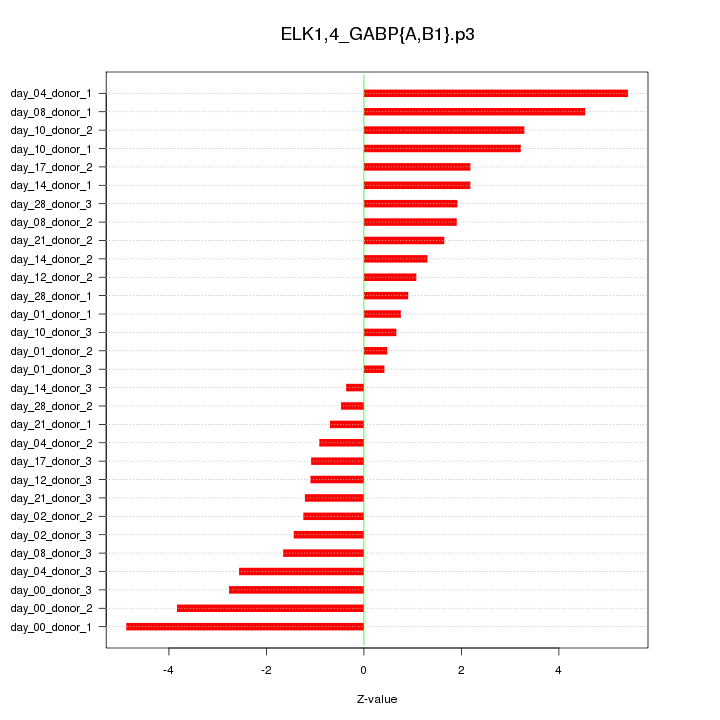
|
chr16_+_80574853
|
10.910
|
NM_130897
|
DYNLRB2
|
dynein, light chain, roadblock-type 2
|
|
chr15_+_71184781
|
6.158
|
NM_001199017
NM_017691
NM_001199018
|
LRRC49
|
leucine rich repeat containing 49
|
|
chr1_-_67390413
|
5.858
|
NM_024763
NM_207014
|
WDR78
|
WD repeat domain 78
|
|
chr17_+_45908992
|
5.470
|
NM_033413
|
LRRC46
|
leucine rich repeat containing 46
|
|
chr11_-_47736896
|
4.856
|
NM_024783
|
AGBL2
|
ATP/GTP binding protein-like 2
|
|
chr11_-_111944757
|
4.842
|
NM_001082619
NM_138789
|
PIH1D2
|
PIH1 domain containing 2
|
|
chr11_+_61129831
|
4.790
|
|
TMEM138
|
transmembrane protein 138
|
|
chr3_-_47324255
|
4.463
|
NM_001134878
NM_022342
NM_182902
|
KIF9
|
kinesin family member 9
|
|
chr11_+_6502607
|
4.389
|
NM_012192
|
FXC1
|
fracture callus 1 homolog (rat)
|
|
chr21_-_33984865
|
4.288
|
NM_021254
|
C21orf59
|
chromosome 21 open reading frame 59
|
|
chr1_+_118472364
|
4.197
|
NM_006784
|
WDR3
|
WD repeat domain 3
|
|
chr20_+_49575347
|
3.956
|
NM_014484
|
MOCS3
|
molybdenum cofactor synthesis 3
|
|
chr3_-_180397261
|
3.819
|
NM_181426
|
CCDC39
|
coiled-coil domain containing 39
|
|
chr1_+_20512577
|
3.783
|
NM_152376
|
UBXN10
|
UBX domain protein 10
|
|
chr11_+_61129407
|
3.779
|
NM_016464
|
TMEM138
|
transmembrane protein 138
|
|
chr2_-_27603584
|
3.747
|
NM_144631
|
ZNF513
|
zinc finger protein 513
|
|
chr2_+_219536719
|
3.706
|
NM_001243313
NM_015690
|
STK36
|
serine/threonine kinase 36
|
|
chr2_-_99771186
|
3.620
|
NM_025244
|
TSGA10
|
testis specific, 10
|
|
chr2_-_27603249
|
3.602
|
NM_001201459
|
ZNF513
|
zinc finger protein 513
|
|
chrX_-_99665270
|
3.585
|
NM_001105243
NM_001184880
NM_020766
|
PCDH19
|
protocadherin 19
|
|
chr2_-_239148545
|
3.558
|
|
HES6
|
hairy and enhancer of split 6 (Drosophila)
|
|
chr10_-_53459309
|
3.459
|
NM_015235
|
CSTF2T
|
cleavage stimulation factor, 3' pre-RNA, subunit 2, 64kDa, tau variant
|
|
chr9_-_100395553
|
3.456
|
|
TSTD2
|
thiosulfate sulfurtransferase (rhodanese)-like domain containing 2
|
|
chr7_+_102105459
|
3.445
|
|
LRWD1
|
leucine-rich repeats and WD repeat domain containing 1
|
|
chr9_-_35749209
|
3.410
|
NM_020944
|
GBA2
|
glucosidase, beta (bile acid) 2
|
|
chr5_-_180018465
|
3.410
|
NM_052863
|
SCGB3A1
|
secretoglobin, family 3A, member 1
|
|
chr16_+_2880172
|
3.345
|
NM_145252
|
ZG16B
|
zymogen granule protein 16 homolog B (rat)
|
|
chr9_-_35749144
|
3.337
|
|
GBA2
|
glucosidase, beta (bile acid) 2
|
|
chr2_-_220110116
|
3.329
|
NM_024506
|
GLB1L
|
galactosidase, beta 1-like
|
|
chr3_-_93781659
|
3.328
|
NM_176815
|
DHFRL1
|
dihydrofolate reductase-like 1
|
|
chr2_-_239148626
|
3.318
|
NM_001142853
NM_018645
|
HES6
|
hairy and enhancer of split 6 (Drosophila)
|
|
chr3_-_127842566
|
3.254
|
NM_003707
|
RUVBL1
|
RuvB-like 1 (E. coli)
|
|
chr20_+_56725982
|
3.183
|
NM_178456
|
C20orf85
|
chromosome 20 open reading frame 85
|
|
chr21_-_33984657
|
3.170
|
|
C21orf59
|
chromosome 21 open reading frame 59
|
|
chr17_-_19281494
|
3.152
|
NM_001243475
|
B9D1
|
B9 protein domain 1
|
|
chrY_+_2709596
|
3.145
|
NM_001008
|
RPS4Y1
|
ribosomal protein S4, Y-linked 1
|
|
chrY_+_2709630
|
3.117
|
|
RPS4Y1
|
ribosomal protein S4, Y-linked 1
|
|
chr1_-_169336999
|
3.031
|
NM_013330
NM_197972
|
NME7
|
non-metastatic cells 7, protein expressed in (nucleoside-diphosphate kinase)
|
|
chr9_-_100395961
|
3.014
|
NM_139246
|
TSTD2
|
thiosulfate sulfurtransferase (rhodanese)-like domain containing 2
|
|
chr19_-_12807394
|
2.989
|
NM_032301
|
FBXW9
|
F-box and WD repeat domain containing 9
|
|
chr9_+_131218402
|
2.959
|
|
ODF2
|
outer dense fiber of sperm tails 2
|
|
chr9_+_131218426
|
2.920
|
|
ODF2
|
outer dense fiber of sperm tails 2
|
|
chr1_-_160001529
|
2.907
|
|
PIGM
|
phosphatidylinositol glycan anchor biosynthesis, class M
|
|
chr3_-_142297565
|
2.885
|
|
ATR
|
ataxia telangiectasia and Rad3 related
|
|
chr17_+_7487125
|
2.869
|
|
MPDU1
|
mannose-P-dolichol utilization defect 1
|
|
chr14_+_100842728
|
2.829
|
NM_001161476
NM_024515
|
WDR25
|
WD repeat domain 25
|
|
chr1_-_47779651
|
2.819
|
NM_001048166
NM_003035
|
STIL
|
SCL/TAL1 interrupting locus
|
|
chr10_+_22634373
|
2.813
|
NM_001253854
NM_001253855
NM_012443
NM_172242
|
SPAG6
|
sperm associated antigen 6
|
|
chr3_+_32726659
|
2.812
|
NM_015442
|
CNOT10
|
CCR4-NOT transcription complex, subunit 10
|
|
chr5_-_35938736
|
2.800
|
NM_001042625
NM_144647
|
CAPSL
|
calcyphosine-like
|
|
chr11_+_61159818
|
2.783
|
NM_001173990
NM_001173991
NM_016499
|
TMEM216
|
transmembrane protein 216
|
|
chr19_-_55791490
|
2.780
|
|
HSPBP1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1
|
|
chr9_+_131218678
|
2.770
|
NM_153433
NM_153440
|
ODF2
|
outer dense fiber of sperm tails 2
|
|
chr9_+_35732629
|
2.748
|
|
CREB3
|
cAMP responsive element binding protein 3
|
|
chr9_-_35748875
|
2.698
|
|
GBA2
|
glucosidase, beta (bile acid) 2
|
|
chr3_-_194392938
|
2.663
|
|
LSG1
|
large subunit GTPase 1 homolog (S. cerevisiae)
|
|
chr11_+_61197574
|
2.633
|
NM_017841
|
SDHAF2
|
succinate dehydrogenase complex assembly factor 2
|
|
chr9_+_131218435
|
2.574
|
|
ODF2
|
outer dense fiber of sperm tails 2
|
|
chr19_+_32896515
|
2.573
|
NM_001172774
|
DPY19L3
|
dpy-19-like 3 (C. elegans)
|
|
chr3_-_129118241
|
2.565
|
|
RPL32P3
|
ribosomal protein L32 pseudogene 3
|
|
chr16_+_57769667
|
2.552
|
|
KATNB1
|
katanin p80 (WD repeat containing) subunit B 1
|
|
chr17_+_4981754
|
2.532
|
|
ZFP3
|
zinc finger protein 3 homolog (mouse)
|
|
chr14_+_105953531
|
2.531
|
|
CRIP1
|
cysteine-rich protein 1 (intestinal)
|
|
chr11_+_61160098
|
2.523
|
|
TMEM216
|
transmembrane protein 216
|
|
chr3_-_142297650
|
2.519
|
NM_001184
|
ATR
|
ataxia telangiectasia and Rad3 related
|
|
chr19_-_55791446
|
2.500
|
|
HSPBP1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1
|
|
chr12_-_105629790
|
2.500
|
NM_001251904
NM_018171
|
APPL2
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2
|
|
chr7_+_39605871
|
2.496
|
NM_020192
|
YAE1D1
|
Yae1 domain containing 1
|
|
chr3_-_93782066
|
2.494
|
NM_001195643
|
DHFRL1
|
dihydrofolate reductase-like 1
|
|
chr11_-_6502563
|
2.493
|
NM_001242854
NM_001242855
NM_001242856
NM_012402
|
ARFIP2
|
ADP-ribosylation factor interacting protein 2
|
|
chr6_+_7390012
|
2.482
|
NM_031480
|
RIOK1
|
RIO kinase 1 (yeast)
|
|
chr11_-_6502524
|
2.479
|
|
ARFIP2
|
ADP-ribosylation factor interacting protein 2
|
|
chr6_+_33422319
|
2.468
|
NM_152735
|
ZBTB9
|
zinc finger and BTB domain containing 9
|
|
chr11_+_71791376
|
2.467
|
NM_001145307
NM_001145308
NM_001205138
NM_145309
|
LRTOMT
|
leucine rich transmembrane and 0-methyltransferase domain containing
|
|
chr16_+_89724175
|
2.451
|
NM_153025
|
C16orf55
|
chromosome 16 open reading frame 55
|
|
chr9_+_131218279
|
2.444
|
NM_001242352
NM_001242353
NM_001242354
NM_002540
NM_153432
NM_153435
NM_153439
|
ODF2
|
outer dense fiber of sperm tails 2
|
|
chr17_-_74137370
|
2.398
|
NM_001454
|
FOXJ1
|
forkhead box J1
|
|
chr7_+_48075107
|
2.387
|
NM_001100159
|
C7orf57
|
chromosome 7 open reading frame 57
|
|
chr16_-_54319966
|
2.369
|
|
IRX3
|
iroquois homeobox 3
|
|
chr19_-_44860798
|
2.365
|
NM_001083335
NM_013380
|
ZFP112
|
zinc finger protein 112 homolog (mouse)
|
|
chr19_-_55791428
|
2.362
|
|
HSPBP1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1
|
|
chr19_-_55791727
|
2.361
|
NM_001130106
NM_012267
|
HSPBP1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1
|
|
chr12_+_124118312
|
2.350
|
NM_001516
|
GTF2H3
|
general transcription factor IIH, polypeptide 3, 34kDa
|
|
chr15_+_44092782
|
2.331
|
|
C15orf63
|
chromosome 15 open reading frame 63
|
|
chr1_+_22351984
|
2.309
|
|
LINC00339
|
long intergenic non-protein coding RNA 339
|
|
chr2_+_172378856
|
2.304
|
NM_001127383
NM_024843
|
CYBRD1
|
cytochrome b reductase 1
|
|
chr19_-_19774472
|
2.303
|
|
ATP13A1
|
ATPase type 13A1
|
|
chr7_+_102987970
|
2.261
|
NM_001204453
NM_002803
|
PSMC2
|
proteasome (prosome, macropain) 26S subunit, ATPase, 2
|
|
chr15_+_44092618
|
2.239
|
NM_001199885
NM_016400
|
C15orf63
|
chromosome 15 open reading frame 63
|
|
chr1_+_44679111
|
2.231
|
NM_001034023
NM_001034024
NM_019100
|
DMAP1
|
DNA methyltransferase 1 associated protein 1
|
|
chr3_-_194392881
|
2.205
|
|
LSG1
|
large subunit GTPase 1 homolog (S. cerevisiae)
|
|
chr2_+_27805892
|
2.191
|
NM_032434
|
ZNF512
|
zinc finger protein 512
|
|
chr19_-_48825150
|
2.183
|
|
CCDC114
|
coiled-coil domain containing 114
|
|
chrY_-_2368579
|
2.173
|
NM_001171135
|
ZBED1
|
zinc finger, BED-type containing 1
|
|
chr13_-_29292961
|
2.172
|
NM_001135919
NM_181785
|
SLC46A3
|
solute carrier family 46, member 3
|
|
chr21_-_46340776
|
2.162
|
|
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit)
|
|
chr11_+_35684418
|
2.139
|
|
TRIM44
|
tripartite motif containing 44
|
|
chr19_-_19774501
|
2.129
|
NM_020410
|
ATP13A1
|
ATPase type 13A1
|
|
chr2_-_28113211
|
2.103
|
NM_022128
|
RBKS
|
ribokinase
|
|
chr14_+_77924367
|
2.097
|
NM_012111
|
AHSA1
|
AHA1, activator of heat shock 90kDa protein ATPase homolog 1 (yeast)
|
|
chr1_+_46049659
|
2.095
|
NM_001195193
NM_002482
NM_152298
|
NASP
|
nuclear autoantigenic sperm protein (histone-binding)
|
|
chr19_+_44645728
|
2.085
|
|
ZNF234
|
zinc finger protein 234
|
|
chr6_+_18155535
|
2.071
|
NM_153042
|
KDM1B
|
lysine (K)-specific demethylase 1B
|
|
chr21_-_46340821
|
2.067
|
|
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit)
|
|
chr17_-_3867585
|
2.066
|
NM_005173
NM_174953
NM_174954
NM_174955
NM_174956
NM_174957
NM_174958
|
ATP2A3
|
ATPase, Ca++ transporting, ubiquitous
|
|
chr17_+_34891340
|
2.063
|
NM_178517
|
PIGW
|
phosphatidylinositol glycan anchor biosynthesis, class W
|
|
chr12_-_25348031
|
2.060
|
NM_001082972
NM_001082973
NM_001204101
NM_001204102
NM_018272
|
CASC1
|
cancer susceptibility candidate 1
|
|
chr2_-_73964427
|
2.050
|
NM_016058
|
TPRKB
|
TP53RK binding protein
|
|
chr17_+_4981721
|
2.030
|
NM_153018
|
ZFP3
|
zinc finger protein 3 homolog (mouse)
|
|
chr14_+_77924415
|
2.030
|
|
AHSA1
|
AHA1, activator of heat shock 90kDa protein ATPase homolog 1 (yeast)
|
|
chr12_+_498513
|
2.024
|
NM_001130147
NM_001130148
|
CCDC77
|
coiled-coil domain containing 77
|
|
chr9_+_35732498
|
2.018
|
|
CREB3
|
cAMP responsive element binding protein 3
|
|
chr10_+_99894426
|
2.010
|
|
C10orf28
|
chromosome 10 open reading frame 28
|
|
chr6_+_32811895
|
2.001
|
|
|
|
|
chr2_+_201754049
|
1.998
|
NM_001136039
NM_001142355
NM_021824
|
NIF3L1
|
NIF3 NGG1 interacting factor 3-like 1 (S. cerevisiae)
|
|
chr1_-_160001723
|
1.997
|
NM_145167
|
PIGM
|
phosphatidylinositol glycan anchor biosynthesis, class M
|
|
chr15_-_85197512
|
1.994
|
NM_032856
|
WDR73
|
WD repeat domain 73
|
|
chrX_+_55478537
|
1.990
|
NM_014061
|
MAGEH1
|
melanoma antigen family H, 1
|
|
chr1_+_217804665
|
1.968
|
NM_138796
|
SPATA17
|
spermatogenesis associated 17
|
|
chr1_-_109969058
|
1.967
|
NM_001199772
NM_002790
NM_001199773
NM_001199774
|
PSMA5
|
proteasome (prosome, macropain) subunit, alpha type, 5
|
|
chr11_-_95522685
|
1.961
|
|
FAM76B
|
family with sequence similarity 76, member B
|
|
chr14_-_54423448
|
1.960
|
NM_001202
NM_130850
|
BMP4
|
bone morphogenetic protein 4
|
|
chr17_+_79935493
|
1.957
|
|
ASPSCR1
|
alveolar soft part sarcoma chromosome region, candidate 1
|
|
chr2_-_99758034
|
1.956
|
NM_182911
|
TSGA10
|
testis specific, 10
|
|
chr19_+_36236460
|
1.955
|
NM_172341
|
PSENEN
|
presenilin enhancer 2 homolog (C. elegans)
|
|
chr19_+_58816757
|
1.955
|
|
ERVK3-1
|
endogenous retrovirus group K3, member 1
|
|
chrX_+_47092362
|
1.953
|
|
USP11
|
ubiquitin specific peptidase 11
|
|
chr3_-_72897575
|
1.951
|
|
SHQ1
|
SHQ1 homolog (S. cerevisiae)
|
|
chr9_+_35732207
|
1.950
|
NM_006368
|
CREB3
|
cAMP responsive element binding protein 3
|
|
chr3_-_51533906
|
1.932
|
|
VPRBP
|
Vpr (HIV-1) binding protein
|
|
chr16_+_699277
|
1.919
|
NM_145294
|
WDR90
|
WD repeat domain 90
|
|
chr20_+_48429375
|
1.913
|
|
SLC9A8
|
solute carrier family 9 (sodium/hydrogen exchanger), member 8
|
|
chr22_+_46067677
|
1.912
|
NM_001167621
NM_013236
|
ATXN10
|
ataxin 10
|
|
chr15_-_85197502
|
1.910
|
|
WDR73
|
WD repeat domain 73
|
|
chr9_+_34458810
|
1.910
|
NM_012144
|
DNAI1
|
dynein, axonemal, intermediate chain 1
|
|
chr5_-_68485278
|
1.906
|
|
|
|
|
chr1_+_246729724
|
1.906
|
|
CNST
|
consortin, connexin sorting protein
|
|
chr11_-_64885124
|
1.903
|
NM_014205
|
ZNHIT2
|
zinc finger, HIT-type containing 2
|
|
chr16_+_89724436
|
1.902
|
|
C16orf55
|
chromosome 16 open reading frame 55
|
|
chr9_-_3525973
|
1.900
|
NM_002919
NM_134428
|
RFX3
|
regulatory factor X, 3 (influences HLA class II expression)
|
|
chr3_-_194392913
|
1.899
|
|
LSG1
|
large subunit GTPase 1 homolog (S. cerevisiae)
|
|
chr9_+_100395704
|
1.891
|
NM_002486
|
NCBP1
|
nuclear cap binding protein subunit 1, 80kDa
|
|
chr17_+_40985444
|
1.885
|
|
PSME3
|
proteasome (prosome, macropain) activator subunit 3 (PA28 gamma; Ki)
|
|
chr2_-_99224868
|
1.882
|
NM_001008215
|
COA5
|
cytochrome C oxidase assembly factor 5
|
|
chr21_-_46340874
|
1.865
|
NM_000211
|
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit)
|
|
chr2_+_85980600
|
1.865
|
NM_032827
|
ATOH8
|
atonal homolog 8 (Drosophila)
|
|
chr8_+_67341244
|
1.863
|
NM_015169
|
RRS1
|
RRS1 ribosome biogenesis regulator homolog (S. cerevisiae)
|
|
chr3_-_183273407
|
1.862
|
NM_130446
|
KLHL6
|
kelch-like 6 (Drosophila)
|
|
chr1_+_246729803
|
1.861
|
|
CNST
|
consortin, connexin sorting protein
|
|
chrY_-_2368963
|
1.850
|
NM_145177
NM_001171136
NM_004729
|
DHRSX
ZBED1
|
dehydrogenase/reductase (SDR family) X-linked
zinc finger, BED-type containing 1
|
|
chr1_-_38273823
|
1.849
|
|
YRDC
|
yrdC domain containing (E. coli)
|
|
chr12_+_74931622
|
1.842
|
|
ATXN7L3B
|
ataxin 7-like 3B
|
|
chr17_-_7218665
|
1.840
|
|
GPS2
|
G protein pathway suppressor 2
|
|
chr2_+_99225084
|
1.826
|
|
UNC50
|
unc-50 homolog (C. elegans)
|
|
chr1_+_161123755
|
1.825
|
|
UFC1
|
ubiquitin-fold modifier conjugating enzyme 1
|
|
chr15_-_89089792
|
1.825
|
NM_001144074
NM_017996
|
DET1
|
de-etiolated homolog 1 (Arabidopsis)
|
|
chr17_+_7486964
|
1.825
|
NM_004870
|
MPDU1
|
mannose-P-dolichol utilization defect 1
|
|
chr12_+_124196941
|
1.818
|
|
ATP6V0A2
|
ATPase, H+ transporting, lysosomal V0 subunit a2
|
|
chr16_+_67226027
|
1.815
|
NM_001950
|
E2F4
|
E2F transcription factor 4, p107/p130-binding
|
|
chr1_-_42921923
|
1.815
|
NM_001146192
NM_032257
|
ZMYND12
|
zinc finger, MYND-type containing 12
|
|
chr13_-_31736031
|
1.814
|
NM_006644
|
HSPH1
|
heat shock 105kDa/110kDa protein 1
|
|
chr12_+_74931550
|
1.812
|
NM_001136262
|
ATXN7L3B
|
ataxin 7-like 3B
|
|
chrX_+_47092264
|
1.812
|
NM_004651
|
USP11
|
ubiquitin specific peptidase 11
|
|
chr17_-_7218370
|
1.805
|
NM_004489
|
GPS2
|
G protein pathway suppressor 2
|
|
chr14_+_45431333
|
1.800
|
NM_015091
|
FAM179B
|
family with sequence similarity 179, member B
|
|
chr9_-_117150217
|
1.795
|
|
AKNA
|
AT-hook transcription factor
|
|
chr3_-_194393205
|
1.789
|
NM_018385
|
LSG1
|
large subunit GTPase 1 homolog (S. cerevisiae)
|
|
chr15_+_82555101
|
1.789
|
NM_001008226
|
FAM154B
|
family with sequence similarity 154, member B
|
|
chr11_-_22647329
|
1.788
|
NM_022725
|
FANCF
|
Fanconi anemia, complementation group F
|
|
chr2_-_70418086
|
1.784
|
NM_017880
|
C2orf42
|
chromosome 2 open reading frame 42
|
|
chr22_+_46067892
|
1.775
|
|
ATXN10
|
ataxin 10
|
|
chr16_-_58163181
|
1.772
|
|
C16orf80
|
chromosome 16 open reading frame 80
|
|
chr16_+_4784261
|
1.772
|
NM_139170
|
C16orf71
|
chromosome 16 open reading frame 71
|
|
chr11_+_65479685
|
1.771
|
|
KAT5
|
K(lysine) acetyltransferase 5
|
|
chrX_+_129473812
|
1.770
|
NM_003951
NM_022810
|
SLC25A14
|
solute carrier family 25 (mitochondrial carrier, brain), member 14
|
|
chr4_-_159644439
|
1.766
|
NM_005038
|
PPID
|
peptidylprolyl isomerase D
|
|
chr2_+_99225060
|
1.756
|
|
UNC50
|
unc-50 homolog (C. elegans)
|
|
chr4_+_980775
|
1.748
|
NM_000203
|
IDUA
|
iduronidase, alpha-L-
|
|
chr20_+_62612464
|
1.745
|
|
PRPF6
|
PRP6 pre-mRNA processing factor 6 homolog (S. cerevisiae)
|
|
chr11_+_2421983
|
1.741
|
|
TSSC4
|
tumor suppressing subtransferable candidate 4
|
|
chr2_+_32390902
|
1.731
|
NM_001193513
NM_001193514
NM_001193515
NM_017964
|
SLC30A6
|
solute carrier family 30 (zinc transporter), member 6
|
|
chr8_+_144329105
|
1.727
|
NM_173832
|
ZFP41
|
zinc finger protein 41 homolog (mouse)
|
|
chr11_-_95522640
|
1.724
|
|
FAM76B
|
family with sequence similarity 76, member B
|
|
chr19_-_45681454
|
1.720
|
NM_024108
|
TRAPPC6A
|
trafficking protein particle complex 6A
|
|
chr8_-_104427319
|
1.716
|
|
SLC25A32
|
solute carrier family 25, member 32
|
|
chr6_-_36842751
|
1.716
|
|
PPIL1
|
peptidylprolyl isomerase (cyclophilin)-like 1
|
|
chr10_-_98480174
|
1.713
|
NM_152309
|
PIK3AP1
|
phosphoinositide-3-kinase adaptor protein 1
|
|
chr10_+_99894380
|
1.705
|
NM_014472
|
C10orf28
|
chromosome 10 open reading frame 28
|
|
chrX_+_150565655
|
1.705
|
NM_001017980
|
VMA21
|
VMA21 vacuolar H+-ATPase homolog (S. cerevisiae)
|
|
chr2_-_241500167
|
1.701
|
|
ANKMY1
|
ankyrin repeat and MYND domain containing 1
|
|
chr5_-_130970803
|
1.694
|
|
RAPGEF6
|
Rap guanine nucleotide exchange factor (GEF) 6
|
|
chr11_-_124670283
|
1.693
|
NM_024631
|
C11orf61
|
chromosome 11 open reading frame 61
|
|
chr20_-_44993000
|
1.687
|
NM_015945
NM_173073
NM_173179
|
SLC35C2
|
solute carrier family 35, member C2
|
|
chr8_-_143751322
|
1.682
|
|
JRK
|
jerky homolog (mouse)
|
|
chr17_+_40985416
|
1.680
|
NM_005789
NM_176863
|
PSME3
|
proteasome (prosome, macropain) activator subunit 3 (PA28 gamma; Ki)
|
|
chr11_+_48002063
|
1.678
|
NM_001098503
NM_002843
|
PTPRJ
|
protein tyrosine phosphatase, receptor type, J
|
|
chr2_+_99225041
|
1.676
|
NM_014044
|
UNC50
|
unc-50 homolog (C. elegans)
|
|
chr16_-_58163286
|
1.675
|
NM_013242
|
C16orf80
|
chromosome 16 open reading frame 80
|
|
chr19_+_8455243
|
1.672
|
|
RAB11B
|
RAB11B, member RAS oncogene family
|
|
chr6_-_36842547
|
1.664
|
|
PPIL1
|
peptidylprolyl isomerase (cyclophilin)-like 1
|